Turn Sequencing Data
Into Microbiome Insight
Welcome to the Directed Study in Medical Sciences (Winter 2026). This course is designed to provide a focused, hands-on introduction to microbiome analysis and bioinformatics, with an emphasis on understanding both the theory and practice behind modern microbiome research.
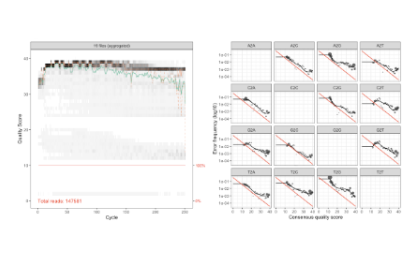
Throughout the course, you will explore core components of microbiome sequencing analysis, including data management, file processing, quality control, and computational approaches used to characterize microbial communities. You will be introduced to different DNA sequencing strategies used in microbiome science and learn how analytical considerations vary across these methods.